Challenges
Please note the latest Challenges Schedule on Apr 30 & May 1
ARE YOU READY FOR A CHALLENGE?! We have SEVEN GRAND CHALLENGES (Please download the flyer) and invite you to JOIN one of our sessions related to your research area! The organization of each challenge is unique. The tasks, metrics, participation requirements, and important dates are given in the links provided below. In general, each challenges is a combination of a contest and a workshop. The contest involves processing ground-truth data and is held during the months preceding the conference. During the conference workshop, additional data may need to be processes, participants will have the opportunity to present their methods, and all attendees will be invited to discuss the results. As the co-chairs of Challenge Sessions, we appreciate the efforts of Challenge organizers and we are excited to be part of ISBI 2014! Please check the links provided by the organizers below for more details and don’t hesitate to contact us for any inquiries. Looking forward to seeing you in Beijing!
| Gultekin Gulsen, PhD e-mail: ggulsen@uci.edu University of California, Irvine,CA, USA |
Jun Zhao, PhD e-mail: junzhao@sjtu.edu.cn Shanghai Jiao Tong University, Shanghai, China |
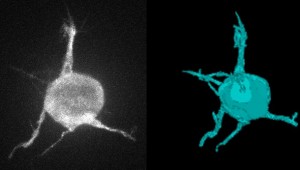
1. Deconvolution – Making the Most of Fluorescence Microscopy
Deconvolution is one of the most common image-reconstruction tasks that arise in 3D fluorescence microscopy. The aim of this challenge is to benchmark existing deconvolution algorithms and to stimulate the community to look for novel, global and practical approaches to this problem.
The challenge will be divided into two stages: a training phase and a competition (testing) phase. It will primarily be based on realistic-looking synthetic data sets representing various sub-cellular structures. In addition it will rely on a number of common and advanced performance metrics to objectively assess the quality of the results.
2. Cell Tracking Challenge 2014
Tracking moving cells in time-lapse video sequences is a challenging task, required for many applications in both scientific and industrial settings. Properly characterizing how cells move as they interact with their surrounding environment is key to understanding the mechanobiology of cell migration and its multiple implications in both normal tissue development and many diseases. In this challenge we will objectively compare and evaluate state-of-the-art whole-cell and nucleus tracking methods using both real (2D and 3D) time-lapse microscopy videos of fluorescently labeled cells and nuclei, along with computer generated video sequences simulating nuclei moving in realistic environments.
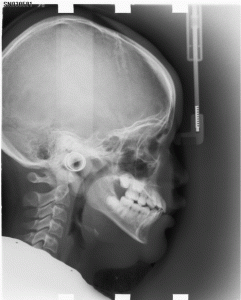
3. Automatic Cephalometric X-Ray Landmark Detection Challenge 2014
Cephalometric analysis is mandatory for the orthodontic analysis and treatment plan making, and marking of critical landmarks in cephalometric analysis is necessary as it provides the interpretation of patients’ bony structures and gives the whole pictures for surgeons in orthognathic surgery (OGS). In clinical practice, manual marking is commonly conducted, but this is very time consuming and subjective. Automated landmark detection of cephalometry could be the solution to facilitate these issues. However, automated landmark detection with high precision and success rate is challenging. From literature review, in recent 7 years the success rate of automatic landmarks has increased from 71% to only 89.5% with accuracy of less than 2.0 mm, which is the accepted accuracy of a landmark in clinical practice.
The goal of Automatic Cephalometric X-Ray Landmark Detection Challenge 2014 is to investigate a suitable automatic landmark detection technology for cephalometric x-ray images and provide a standard evaluation framework with a clinical dataset. The challenge participating teams (two co-authors in each participating team) will be invited to contribute to a joint journal paper describing and summarizing the challenge outcome, which will be submitted to a high-impact SCI journal in the field.
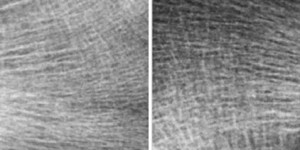
4. Bone Texture Characterization
Osteoporosis is defined as a skeletal disorder characterized by compromised bone strength predisposing to an increased risk of fracture. The most common method for osteoporosis diagnosis is to estimate Bone Mineral Density (BMD) by dual-energy X-ray absorptiometry. However, BMD alone represents only 60% of fracture prediction. The characterization of trabecular bone microarchitecture has been recognized as an important factor and completes the osteoporosis diagnosis using BMD, but it cannot be routinely obtained by noninvasive methods and requires a bone biopsy with histomorphometric analysis.The evaluation of osteoporotic disease from bone radiograph images presents a major challenge for pattern recognition and medical applications. Textured images from the bone microarchitecture of osteoporotic and healthy subjects show a high degree of similarity, thus drastically increasing the difficulty of classifying such textures.
The goal of this Challenge is to identify osteoporotic cases from healthy controls on 2D bone radiograph images, using texture analysis. It consists of two populations composed of 87 control subjects (CT) and 87 patients with osteoporotic fractures (OP). The Challenge is opened to academia and industry. Already published methods can also be submitted. Some of the participants will be invited to present their work at the workshop during a half day session (28 April ¨C 2 May, 2014) in the form of an oral or poster presentation. The final results from this challenge will be summarized and submitted to a journal paper, presenting and comparing all used texture analysis methods used to achieve osteoporosis diagnosis on bone radiograph images. The journal paper will be written and co-authored by the organizers and the selected participants.
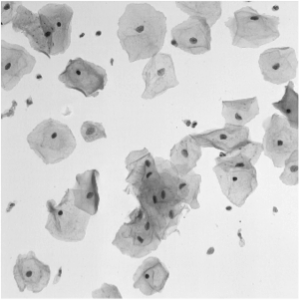
5. Overlapping Cervical Cytology Image Segmentation Challenge
The automated detection and segmentation of overlapping cells using microscopic images obtained from Pap smear can be considered to be one of the major hurdles for a robust automatic analysis of cervical cells. The Pap smear is a screening test used to detect pre-cancerous and cancerous processes, which consists of a sample of cells collected from the cervix that are smeared onto a glass slide and further examined under a microscope. The main factors affecting the sensitivity of the Pap smear test are the number of cells sampled, the overlap among these cells, the poor contrast of the cell cytoplasm, and the presence of mucus, blood and inflammatory cells. Automated slide analysis techniques attempt to improve both sensitivity and specificity by automatically detecting, segmenting and classifying the cells present on a slide.
In this challenge, the targets are to extract the boundaries of individual cytoplasm and nucleus from overlapping cervical cytology images.
6. VISCERAL Organ Segmentation and Landmark Detection Benchmark
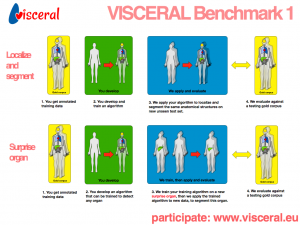 While a growing number of benchmark studies compare the performance of algorithms for automated organ segmentation or lesion detection in images with restricted field of views, no efforts have been made so far towards benchmarking these and related routines for the automated identification and segmentation of bones, inner organs and relevant substructures visible in images with wide-field of view, showing all of the abdomen, the trunk, or even the whole body. In this benchmark we will evaluate related segmentation and detection algorithms on a large dataset of clinical wide-field-of-view MRI and CT scans – from abdomen, trunk and whole body – in which major organs and their substructure have been manually delineated, as well as specific anatomical interest points or landmarks.
While a growing number of benchmark studies compare the performance of algorithms for automated organ segmentation or lesion detection in images with restricted field of views, no efforts have been made so far towards benchmarking these and related routines for the automated identification and segmentation of bones, inner organs and relevant substructures visible in images with wide-field of view, showing all of the abdomen, the trunk, or even the whole body. In this benchmark we will evaluate related segmentation and detection algorithms on a large dataset of clinical wide-field-of-view MRI and CT scans – from abdomen, trunk and whole body – in which major organs and their substructure have been manually delineated, as well as specific anatomical interest points or landmarks.
Our data set comprises about 300 training volumes and 100 test volumes with annotated anatomical structures, such as kidney, spleen, liver, lung, urinary bladder, pancreas, adrenal gland, and others, as well as anatomical landmarks, such as the lateral end of the clavicula, crista iliaca, symphysis, trochanter major/minor, tip of aortic arch, trachea bifurcation, aortic bifurcation, crista iliaca. The benchmark will take place in a cloud environment provided by the organizers.
7. Pancreas Segmentation from 3D Abdominal CT images
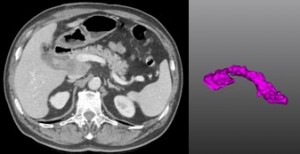 Computerized tomography (CT) represents the commonest method for pancreas image. The segmentation of pancreas tissue from CT is highly applied for a variety of clinical applications since it is fundamental for further pancreas image processing such as pancreas lesion detection and pancreatic cancer analysis. However, it is challenging to achieve the accurate segmentation of pancreas tissue. The intensity of pancreas is similar with that of the peripheral zone around it, which easily leads to under or oversegmentation at weak boundary. In addition, distortion and noise often impact computerized tomography images, which increases difficulties to gain correct segmentation of pancreas.
Computerized tomography (CT) represents the commonest method for pancreas image. The segmentation of pancreas tissue from CT is highly applied for a variety of clinical applications since it is fundamental for further pancreas image processing such as pancreas lesion detection and pancreatic cancer analysis. However, it is challenging to achieve the accurate segmentation of pancreas tissue. The intensity of pancreas is similar with that of the peripheral zone around it, which easily leads to under or oversegmentation at weak boundary. In addition, distortion and noise often impact computerized tomography images, which increases difficulties to gain correct segmentation of pancreas.
The aim of this challenge is to benchmark existing 3D pancreas segmentation methods from computer tomography and to inspire the participants to work out novel and practical approaches to identify the connected tissue and the pancreas. 30 CT data sets will be provided to participants for segmentation. 30 CT data sets will be provided with expert manual segmentation for algorithm training. The researchers are welcomed to submit their unpublished and already published methods to the Challenge. Some of submitters will be invited to present their work at the workshop in ISBI 2014 in the form of a poster or oral presentation. The final results and the corresponding methods on the Challenge will be compared, summarized and written into a paper, which will be submitted to a journal in the field. The journal paper will be finished and co-authored by the organizers and the authors of the selected teams.